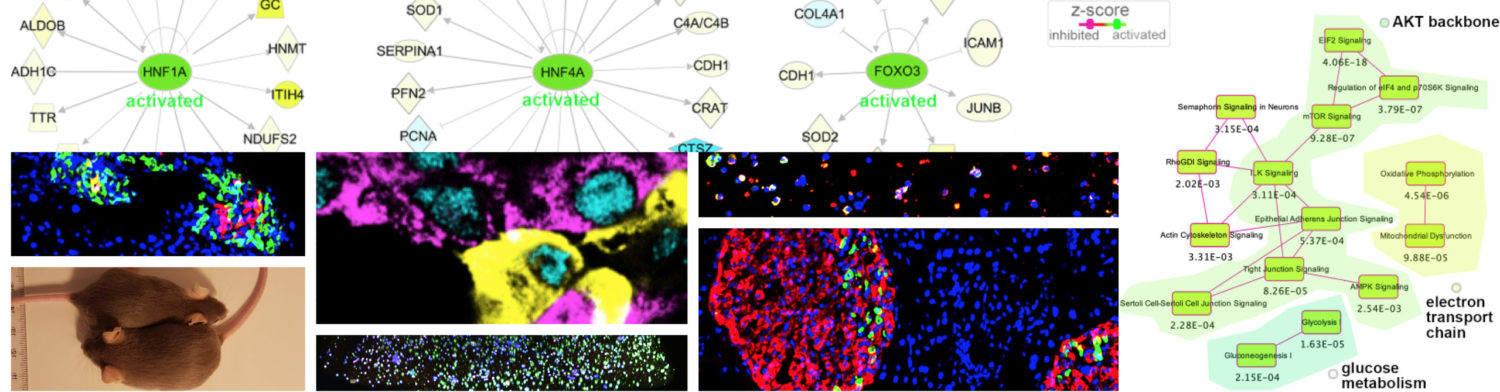
For the past five years, our collaborator Helge Ræder and us showed that human induced pluripotent stem cells (hiPSC) provide an excellent tool for disease modelling. However, in vitro differentiation is highly sensitive to culture conditions. By re-creating a spatial (3D) environment in the dish we argue that it will facilitate phenotype detection for certain mutations. Ræder’s group focused on studying the effect of mutation in HNF4A by using size-adjusted cell aggregates and alginate capsules. The results of this study were published today, showing that:
- 3D context was critical to facilitate the detection of mutation-specific phenotypes;
- In 3D cell aggregates: irregular cell clusters were observed, and lower levels of structural proteins were identified by proteome analysis;
- in 3D alginate capsules: altered levels of glycolytic proteins in the glucose sensing apparatus were detected.
Continue your reading here:
Authors:
Diabetes 2022 14 January 2022
DOI information: 10.2337/db20-1279