This week’s journal club focuses on a study from BMC Genomics (2024) using single-cell sequencing datasets to characterize the transcriptional identity of islets from in vitro differentiation, fetal islets, and adult islets.
by Agnes Sandvik
Aim
In the introduction the authors point out that that even though several protocols on how to generate SC-ϐ cells in vitro do exist, they differ in media composition and culture systems and consistently produce heterogenous 3D cell clusters containing off-target cell types that are transcriptionally and functionally immature.
They state that improving the existing protocols requires a better understanding of the gene expressing programs that define true ϐ-cell identity and regulate differentiation and maturation. They point to single-cell RNA sequencing as a tool that has helped reveal off-target populations and changes in identity and maturation after transplantation, but that no study has systematically compared SC-islets produced by the different protocols or benchmarked them against fetal and adult human islets.
The aim of the study is thereby to address this knowledge gap and determine how well SC-ϐ cells mimic normal human ϐ-cell development.
Methods:
To determine the transcriptional identity of the cells they used published scRNA-seq datasets of SC-islets from multiple protocols, both before and after transplantation, and datasets from human adult and fetal islets. They then performed statistical and comparative analysis of transcription profiles across different β-cell maturation states.
Results:
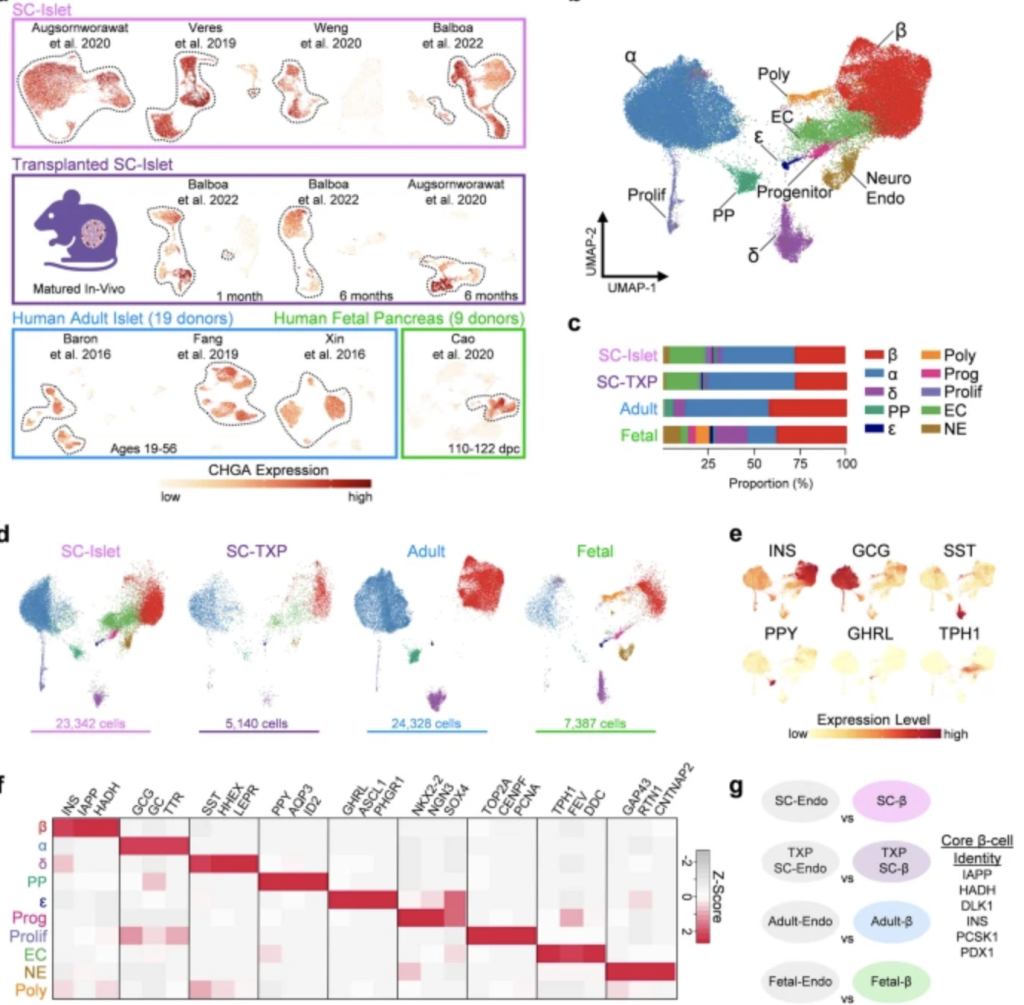
Identification of pancreatic endocrine cell types using integrated transcriptomic atlas
Quality control was used to filter out dead cells and duplicates before they isolated chromogranin A positive (CHGA+) cells from; SC-islets cultured to their endpoint, SC-islets transplanted into kidney capsules of mice and primary fetal and adult islets (Fig 1a) as probable endocrine cell-types and performed unsupervised clustering. This revealed ten distinct endocrine populations (Fig 1b). The adult islets only had six of the identified cell populations (ϐ, α, PP, δ, ε and proliferating endocrine), these were also present in fetal- and SC-islets (Fig 1c-d).
The identification and validation of these endocrine cell populations were further assisted by high expression levels of hormones INS, GCG, SST, PPY and GHRL, along with enrichment of other cell-specific markers (Fig 1e-f). They identified clusters of cells consistent with previous findings as well as one interesting finding of a population of cells with neuroendocrine (NE) features, marked by enrichment of GAP43, RTN1 and CNTNAP2 that was present in both SC-islets and fetal islets. The identity and role of these cells with neuronal properties in the developing human islet has not been characterized previously, suggesting that their generated dataset can be utilized for more precise identification of islet endocrine cell types than from the individual clustering of SC-islet scRNA-seq datasets.
Genes enriched in ϐ-cells across all tissue sources compared to other endocrine cells were also identified for an attempt to suggest a universal definition of ϐ-cell identity (Fig 1g).
Directed differentiation protocols produce transcriptionally similar SC-islets
To investigate commonalities and differences between transcriptional profiles of SC-islets produced by different protocols SC- adult- and fetal-ϐ cells were isolated from the combined dataset and re-clustered (Fig 2a). Based on Pearson correlation analysis they found that SC-ϐ cells from all protocols appeared transcriptionally similar in comparison to adult and fetal ϐ-cells (Fig 2b) and expressed significantly less G6PC2, IAPP, HADH, UNC3, CHGB, ADCYAP1 and SIX3 than adult ϐ-cells (Fig 2c).
They also identified some unique transcriptional profiles from each differentiation protocol (Fig 2d), but state that further investigation is needed to determine if these differences are important for SC-ϐ cell function. They point out that Augsornworawat and Veres used Hues8 hESC and Balboa and Weng used H1 hESC for their differentiation protocol, but state that it is unclear whether the observed differences is due to genetic backgrounds of the utilized cell lines or experimental procedures.
A similar comparative analysis for SC-EC cells from each SC-islet dataset, marked by high expression of TRPH1 through isolation and re-clustering SC-EC cells was also performed (Fig 2e). Expression of key SC-EC marker genes and Pearson correlation suggested that overall transcriptional profile of SC-EC was similar across protocols (Fig 2f-g). When they analyzed differentially expressed genes (DEGs) they saw key differences between the SC-EC cells from the different protocols (Fig 2h). Notably, cells derived by Augsornworawat, et al. had increased expression of α-cell markers and SC-EC cells generated by Balboa, et al. were unique for having high expression of ribosomal genes, like the SC-β cells from this study.
SC-ϐ cells are transcriptionally more mature than fetal ϐ-cells
Previous studies have shown that SC-ϐ cells are transcriptionally immature. Here they characterized the maturation by comparing the global transcription landscape to adult and fetal ϐ-cells (Fig 3a). Through pearson correlation they determined that SC-ϐ cells correlate more with the adult ϐ-cells than what the fetal ϐ-cells does (Fig 3b). However, when they did pairwise comparison between SC-ϐ cells and their primary counterparts they also found DEGs with > 2 fold change enriched in SC-ϐ cells that serve important roles in neurons (Fig 3c). A higher maturity of SC-ϐ cells compared to fetal ϐ-cells were also supported when they compared expression levels of well-known ϐ-cell maturation markers, like INS, and found that while they were present in lower levels in SC-ϐ cells they were mostly non-existent in fetal ϐ-cells (Fig 3e). They also show that fetal ϐ-cells have higher levels of genes important for the exocrine pancreas (Fig 3e-f) and low expression of ribosomal genes, likely necessary for production of peptides (Fig 3g) to further support their findings.
SC-ϐ cells have persistent activity of progenitor transcription factors
The next question they ask is how closely related this immature transcriptional state is to a ϐ-cell progenitor state. They observed that both SC-ϐ and fetal ϐ-cells had significantly higher enrichment of transcription factors than adult ϐ-cells (Fig 4a-b), and that nearly all transcription factors expressed in ϐ-cell progenitor states were expressed in a higher percentage of SC-ϐ cells than adult or fetal ϐ-cells (Fig 4c). Through regulon analysis they further identified the most enriched gene regulatory networks between SC-ϐ, adult- and fetal-ϐ cells (Fig 4d) and found once again that transcription factors associated with ϐ-cell progenitor states were most active in SC-ϐ cells (Fig 4e-g),
Dysregulated transcription factor activity drives neuronal gene program in SC-ϐ cells
Finally, they wanted to find the major gene programs enriched in SC-ϐ cells that account for their transcriptional immaturity. They did gene set enrichment analysis (GSEA) between SC-ϐ cells and adult ϐ-cells and found that the SC-ϐ cells had increased activity in genes related to neuronal morphology and function (Fig 5a-b). They point out that ϐ-cells and neurons have been shown to share a variety of similarities, including exocytosis of insulin-containing granules, which are controlled by many genes with a similar role in neurotransmission. They thereby studied the expression of components in the insulin exocytosis machinery but saw no difference between SC and adult ϐ-cells (Fig 5c). Through further investigation of neuronal traits in SC-ϐ cells they found overexpression of genes encoding neurofilaments and proteins involved in axon guidance, neuronal migration, neurotransmission and action potential (Fig 5d). They also produced their own SC-islets and compared them to cadaveric human islets to confirm that they expressed neuronal genes at a significantly higher level than the adult islets (Fig 5e).
They found that the transcription factors shared in pancreatic and neuronal development had a higher expression in SC-ϐ than adult and fetal ϐ-cells, especially genes of interest (Fig 6a). To test if persistent activity of progenitor associated transcription factors activate neuronal gene programs in SC-ϐ cells they analyzed downstream targets of shared transcription factors in pancreatic and neuronal development and found many factors with gene targets involved in neuronal gene programs that were highly active in SC-ϐ cells, but not present in adult ϐ-cells (Fig 6b). Lastly, they cross checked a list of genes defining the neuronal program enriched in SC-ϐ cells with target genes of active transcription factors in adult ϐ-cells, and found that nearly all transcription factors that are predicted to control the most neuronal target genes were enriched in SC-ϐ cells in comparison to adult ϐ-cells (Fig 6c).
Throughout the article they also point to supplementary experiments of transplantation into the kidney capsule of mice for 1-6 months where they found that the SC-ϐ cells both gained an increased expression of maturation markers and decreased expression of transcription factors associated with ϐ-progenitor states to more closely mirror the adult ϐ-cells as well as losing their neuronal properties.
Conclusions:
They conclude that they have been able to demonstrate that SC-ϐ cells are transcriptionally more similar to adult than fetal ϐ-cells and provide greater detail for both ϐ-cells and other cell types than what has been done before. They hope this resource will serve as a tool for hypothesis generation in hopes of further optimizing protocols for the generation of SC-ϐ cells.
Some of their main findings:
- The transcriptional identities of all major cell types, including SC-ϐ cells, in general were very similar among all differentiation protocols. They all had low expression of MAFA and UNC3, indicating that development of a protocol that can generate cells expressing high levels of these maturation markers is lacking in the field.
- SC-ϐ cells differ from adult ϐ-cells through expression of neuronal and progenitor transcriptional programs.
- Interestingly SC-EC and SC-β cells, despite being distinct cell types, share commonalities in this irregular neural gene program. This presents the hypothesis that the dysregulated neural transcriptional profile present in SC-β development may contribute to the generation of SC-EC cells.
Continue your reading here:
Mason D Schmidt, Matthew Ishahak, Punn Augsornworawat, Jeffrey R Millman. Comparative and integrative single cell analysis reveals new insights into the transcriptional immaturity of stem cell-derived β cells
BMC Genomics 2024 Jan 24;25(1):105. doi: 10.1186/s12864-024-10013-x.

1 comment for “Journal Club: Comparative and integrative single cell analysis reveals new insights into the transcriptional immaturity of stem cell-derived ϐ-cells”