 My PhD work was focused on the characterization of the molecular and cellular basis of regeneration and homeostasis in one of the best-known regenerative model systems, hydra. My studies were carried out in the Laboratory of regeneration and adult neurogenesis under the supervision of Prof. Brigitte Galliot at the Department of Genetics and Evolution, Faculty of Science, in University of Geneva.
My PhD work was focused on the characterization of the molecular and cellular basis of regeneration and homeostasis in one of the best-known regenerative model systems, hydra. My studies were carried out in the Laboratory of regeneration and adult neurogenesis under the supervision of Prof. Brigitte Galliot at the Department of Genetics and Evolution, Faculty of Science, in University of Geneva.
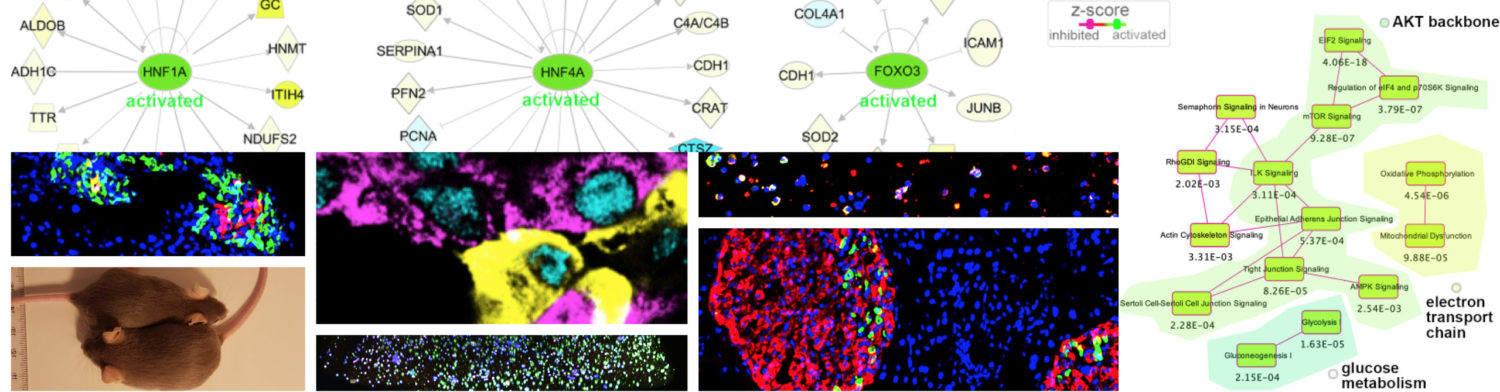
The scarcity of available commercially standardized tools boosted my creativity and stimulated me to establish, develop and implement a series of setups, tools and methods, including a RNA silencing strategy, cloning of novel genes, antibody validation, multiplex detection by RNA FISH, confocal live cell imaging and labeling techniques allowing the monitoring of the regeneration process (Chera et al., 2006 J. Cell Sci., Miljkovic-Licina et al., 2007 Development). As an outcome of this creative investment, I uncovered an unexpected level of complexity governing hydra head-regeneration involving an apoptosis-driven Wnt3a-induced surge of dedifferentiation followed by compensatory proliferation required for an efficient regenerative response. This discovery triggered a paradigm shift, challenging the classical views of hydra regeneration and introducing for the first time in the regenerative field the idea of a dual program of regeneration depending on environmental and internal cues (Chera et al., 2010 Dev. Cell).
Complete publication list related to my PhD work:
- Chera S, Ghila L, Wenger Y, Galliot B. Injury-induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration 2011 Dev Growth Differ. 53(2):186-201 PMID: 21338345
- Galliot B, Chera S. The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration 2010 Trends Cell Biol. 20(9):514-23 PMID: 20691596
- Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu WC, Martinou JC, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration 2009 17(2)279-89 PMID: 19686688
- Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in Hydra: a response to starvation and stress in early animal evolution 2009 Biochim Biophys Acta. 1793(9):1432-43 PMID: 19362111
- Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view 2009 Dev Biol. 332(1), 2-24 PMID: 19465018
- Buzgariu W, Chera S, Galliot B. Methods to investigate autophagy during starvation and regeneration in Hydra 2008 Methods in Enzymology 451, 409-37 PMID: 19185734
- Miljkovic-Licina M, Chera S, Ghila L, Galliot B. Head regeneration in wild-type hydra requires de novo neurogenesis 2007 Development 134:1191-1201 PMID: 17301084
- Galliot B, Miljkovic-Licina M, Ghila L, Chera S. RNAi gene silencing affects cell and developmental plasticity in hydra 2007 C. R. Biol. 330:491-497 PMID: 17631443
- Chera S, Kaloulis K, Galliot B. The cAMP response element binding protein (CREB) as an integrative HUB selector in metazoans: clues from the hydra model system 2007 BioSystems 87:191-203 PMID: 17030409
- Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype 2006 J. Cell Sci. 119:846-857 PMID: 16478786
- Galliot B, Miljkovic-Licina M, de Rosa R, Chera S. Hydra, a niche for cell and developmental plasticity 2006 Sem. Cell Dev. Biol. 17:492-502 PMID: 16807002
- Kaloulis K, Chera S, Hassel M, Gauchat D, Galliot B. Reactivation of developmental programs: the CREB pathway is involved in hydra head regeneration 2004 Natl. Acad. Sci. USA 101:2363-2368 PMID: 14983015
- Gauchat D, Escriva H, Miljkovic-Licina M, Chera S, Langlois MC, Begue A, Laudet V, Galliot B. The orphan COUP-TF nuclear receptors are markers for neurogenesis from cnidarians to vertebrates 2004 Dev. Biol. 275:104-123 PMID: 15464576