This week’s journal club focuses on a study from Drugs in R&D (2024) aiming to assess the activity of trametinib against a panel of tumor cells from patients with AML ex vivo.
Quesnel-Vallières et al. investigate why only a subset of acute myeloid leukemia (AML) samples respond to the MEK inhibitor trametinib, and how molecular features beyond RAS mutations might predict sensitivity. Although trametinib has had limited success in AML, identifying biomarkers to stratify likely responders could improve its clinical utility.
To explore this, the researchers treated a cohort of primary AML blast samples ex vivo with trametinib and measured their sensitivity (IC50). They also performed RNA sequencing on the same samples to correlate gene expression patterns with drug response. They then asked: what transcriptomic features distinguish sensitive vs. resistant AML cells?
They found that about 10 of 37 samples were highly sensitive (low nanomolar IC50), while most were resistant. Interestingly, although RAS mutations were enriched in sensitive samples, RAS status alone was not sufficient to predict sensitivity: some RAS-wildtype patients were also sensitive to trametinib.
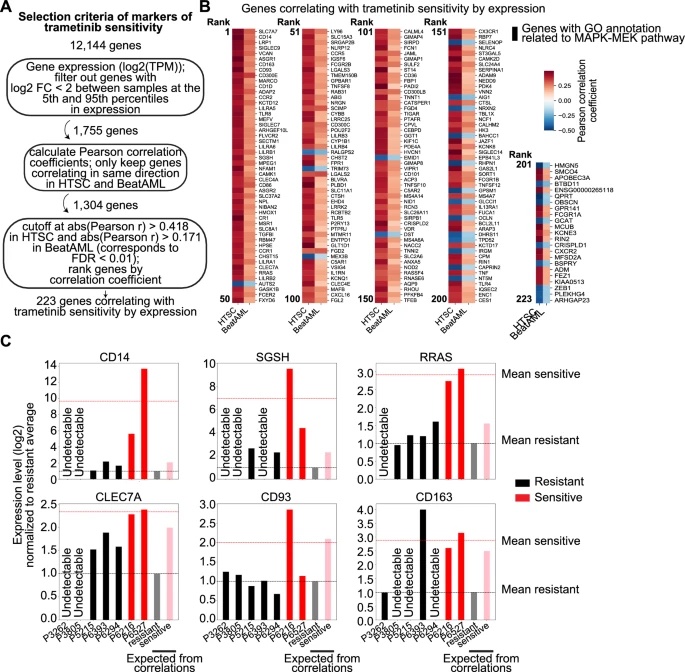
Through correlation and clustering analyses of RNA expression data, the researchers identified a set of ~223 genes whose expression strongly correlated with trametinib sensitivity across both their cohort and an external AML dataset (Beat-AML). Most of these genes were upregulated in sensitive samples. Among these genes, CD14, a marker of monocytic differentiation, stood out for its strong correlation and was validated at the RNA and protein levels in independent AML samples.
Gene set enrichment analysis revealed that the genes positively correlated with trametinib sensitivity were enriched for myeloid/monocyte differentiation signatures. Sensitive AML samples exhibited transcriptomic profiles closely resembling those of mature monocytes, whereas resistant samples more closely resembled immature or progenitor states. In particular, high surface expression of CD14 on blasts was strongly associated with trametinib sensitivity ex vivo.
They propose that AML blasts with more differentiated/monocytic features are more susceptible to MEK inhibition, possibly because of functional ties between differentiation status, MAPK signaling, and responsiveness to trametinib. They further suggest that combining RAS mutation status with surface markers, such as CD14, may help stratify patients who might benefit from trametinib therapy.
In sum, this work demonstrates that canonical oncogenic mutations do not alone determine trametinib sensitivity in AML but are strongly associated with a myeloid differentiation program. Incorporating differentiation marker expression (e.g., CD14) alongside genomic data may improve patient selection for MEK-targeted treatments in AML.
Journal Club: continue your reading here:
Quesnel-Vallières M, Schultz DC, Orlenko A, Lo Y, Moore J, Ritchie M, Roth D, Carroll M, Barash Y, Lynch KW, Cherry S. Trametinib Sensitivity is Defined by a Myeloid Differentiation Profile in Acute Myeloid Leukemia.
Drugs R D. 2024 Sep;24(3):489-499. doi: 10.1007/s40268-024-00491-5. Epub 2024 Sep 24.
PMID: 39316279
