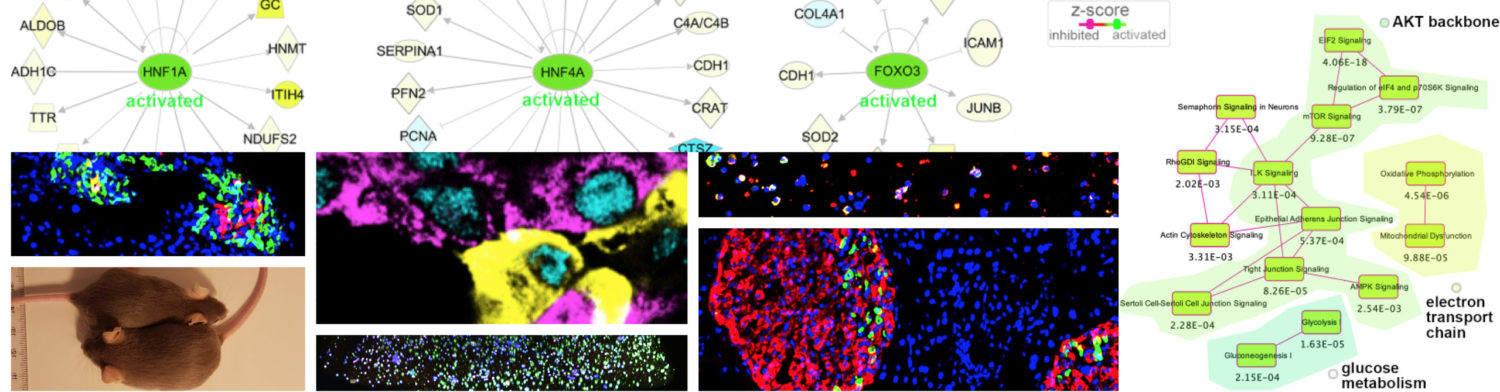
Congratulation to Yngvild for her published article, in which our collaborators show that:
- some reprogrammed cell lines present an unstable state leading to spontaneously differentiate and change their cellular phenotype and colony morphology;
- by comparing the proteome landscapes of 20 reprogrammed cell lines classified as stable and unstable based on long-term colony morphology, distinct proteomic signatures associated with stable colony morphology and with unstable colony morphology were identified;
- the typical pluripotency markers (POU5F1, SOX2) were present with both morphologies;
- epithelial to mesenchymal transition (EMT) protein markers were associated with unstable colony morphology;
- the transforming growth factor beta (TGFB) signalling pathway was predicted as one of the main regulator pathways involved in this process;
- by assessing both spontaneous embryonic body (EB) formation and directed differentiation, the reprogrammed lines with an unstable colony morphology showed reduced differentiation capacity.
To conclude, different defined patterns of colony morphology in reprogrammed cells were associated with distinct proteomic profiles and different outcomes in differentiation capacity.
Continue your reading here:
Authors: Bjørlykke Y, Søviknes AM, Hoareau L, Vethe H, Mathisen AF, Chera S, Vaudel M, Ghila LM, Ræder H. Stem Cells International 2019 Nov 18;2019:8036035. doi: PMID:31827534
DOI information: 10.1155/2019/8036035