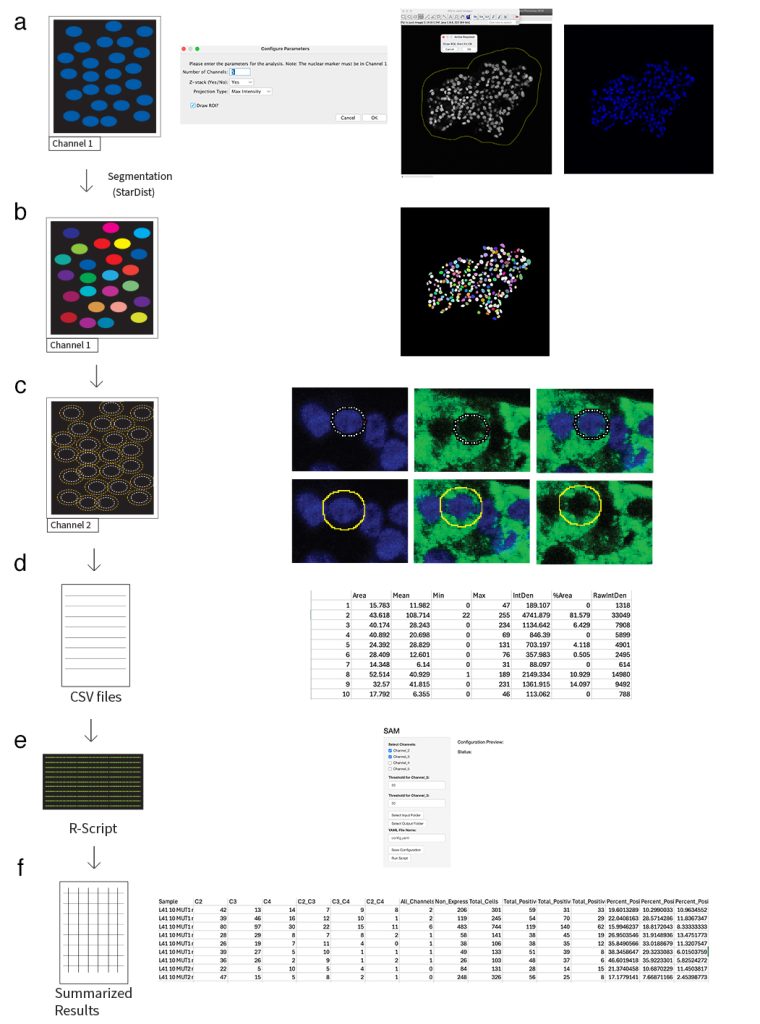
After many months of persuasion, Lucas published his semi-automated quantification tool that he developed over 3 years of quantification of thousands of sc-islets and pancreatic sections. His tool is now available both in GitHUB and as a manuscript (currently as a preprint) to showcase its use and applications.
If you want to test this tool or if you have comments, Lucas is more than happy to adjust and improve this tool. The source code of SAM along with all documentation has been deposited in GitHub and is freely accessible at https://github.com/LucasUnger/SAM
Using this tool, several colleagues from the lab tested different tissues (sc-islets, pancreatic or bladder sections) and compared with other counting methods. Lucas’ SAM proved to be as reliable but with increase control of different parameters. By adding also an user interface, Lucas increased its accessibility. These tests and comparisons are now submitted for publication, and available also in ResearchSquare as preprint:
Continue your reading here:
SAM: A FIJI-Based Tool for Semi-Automated Quantification of Cytoplasmic Expression in Cells
Authors: Unger L, Larsen U, Sharmine S, Hossain MK, Legøy TA, Vaudel M, Ghila L, Chera S.
ResearchSquare
DOI information: 10.21203/rs.3.rs-6466373/v1

1 comment for “PrePrint: SAM: A FIJI-Based Tool for Semi-Automated Quantification of Cytoplasmic Expression in Cells”